Our Research Projects
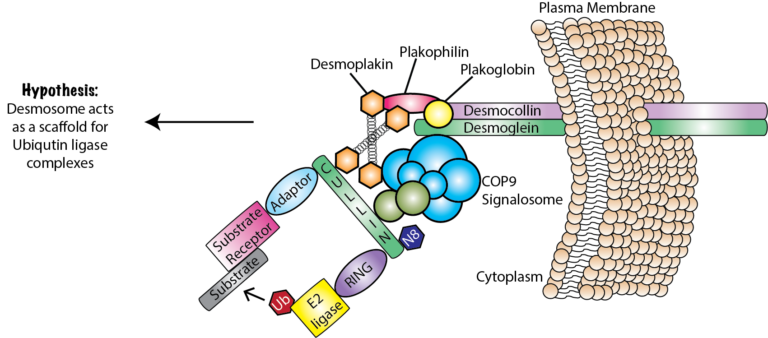
The desmosome is a cell junctional protein complex that links to the intermediate filament cytoskeleton, and is found in simple and stratified epitheliums, such as the epidermis. The skin is composed of self-renewing stem cells that undergo a terminal differentiation program to develop the suprabasal cells of the epidermis. My research identified a role for the desmosomal cadherin, Desmoglein1 (Dsg1), in promoting epidermal differentiation through its interaction with the COP9 signalosome (Najor et al., 2017). One of my main research goals is to continue this project through exploring the complexity of this process specifically in the following two ways:
- How does the desmosome-COP9 signalosome regulate cullin ubiquitin ligases, and to what extent is that required for differentiation.
- How does the desmosome-COP9 signalosome regulate autophagy, and to what extent is that required for differentiation.
Course-Based Undergraduate Research Experience (CURE) Projects
CUREs are designed to provide students with hands-on research experiences, where students develop testable hypotheses that are novel and contribute to the field. They combine the benefit of traditional coursework with real-world application, allowing students to develop better crucial and analytical thinking skills. Students in my CUREs (taken in Cell and Molecular Biology Lab -BIO4750) participate in generating a testable hypothesis, performing the research, collaborating with their peers, analyzing the results, communicating their findings in written and oral format (manuscript drafts, poster sessions and/or lightning talks), and reflecting on their experience.
The role of the desmosome-COP9 signalosome in epithelial cancers- CURE (2016-2019)
I had previously identified the novel interaction between the desmosome and COP9 signalosome in human keratinocytes (Najor et al., 2017), but very little has been done to understand its role in epithelial cancers. This project was designed (1) identify the expression pattern of COP9 components in different human cancer cell lines, (2) identify signaling defects caused by the loss of Cops3 and the loss of Cullin-3. From 2016-2019, students in Cell and Molecular Biology Lab (BIO4750) were presented with the background of the project, and a more global hypothesis. Students were then allowed to further that hypothesis into something more specific based on the reagents that were available. Students used techniques of Western blotting and immunofluorescence to test their hypotheses. Even though students were in the confines of my antibody library, they were still able to participate in novel discovery, which had a broader relevance and impact since the data was used as preliminary results supporting my NIH proposals.
Uncovering genes of unknown functions (ORFans) in the yeast model Saccharomyces cerevisiae CURE (2021-present)
This is an NSF-RCN-UBE funded project, that aims to uncover novel functions of unknown genes through bioinformatics and wet-lab modules. The project is very flexible and there are a variety of ways to execute within a semester. Students in Cell and Molecular Biology Lab (BIO4750) use the bioinformatics modules (available on ORFan Gene Project website) to assist in developing their hypothesis, and then students are required to test that hypothesis. Most students choose to perform fitness tests with different stressors (glycerol, UV, sorbitol, cyclohexamide, temperature shift, ethanol, hydrogen peroxide etc), but they are able to design any experiment they choose. If the student really enjoyed their experience, they are then welcomed to join my lab at the end of the course to continue their project!
